FileSourceScanExec Physical Operator¶
FileSourceScanExec is a DataSourceScanExec that represents a scan over a HadoopFsRelation.
Creating Instance¶
FileSourceScanExec takes the following to be created:
- HadoopFsRelation
- Output Attributes
- Required Schema
- Partition Filters
-
optionalBucketSet -
optionalNumCoalescedBuckets - Data Filters
-
TableIdentifier -
disableBucketedScanflag (default:false)
FileSourceScanExec is created when:
- FileSourceStrategy execution planning strategy is executed (for LogicalRelations over a HadoopFsRelation)
Data Filters¶
FileSourceScanExec is given Data Filters (Expressions) when created.
The Data Filters are data columns of the HadoopFsRelation (that are not partition columns that are part of Partition Filters) in FileSourceStrategy execution planning strategy.
Partition Filters¶
FileSourceScanExec is given Partition Filters (Expressions) when created.
The Partition Filters are the PushedDownFilters (based on the partition columns of the HadoopFsRelation) in FileSourceStrategy execution planning strategy.
Node Name Prefix¶
nodeNamePrefix is always File.
val fileScanExec: FileSourceScanExec = ... // see the example earlier
assert(fileScanExec.nodeNamePrefix == "File")
scala> println(fileScanExec.simpleString)
FileScan csv [id#20,name#21,city#22] Batched: false, Format: CSV, Location: InMemoryFileIndex[file:/Users/jacek/dev/oss/datasets/people.csv], PartitionFilters: [], PushedFilters: [], ReadSchema: struct<id:string,name:string,city:string>
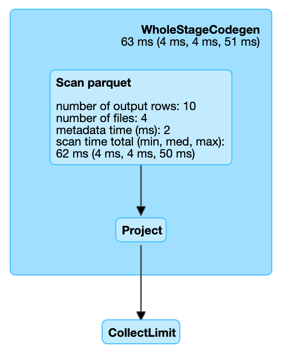
Performance Metrics¶
| Key | Name (in web UI) | Description |
|---|---|---|
| filesSize | size of files read | |
| metadataTime | metadata time (ms) | |
| numFiles | number of files | |
| numOutputRows | number of output rows |
Columnar Scan Metrics¶
The following performance metrics are available only with supportsColumnar enabled.
| Key | Name (in web UI) | Description |
|---|---|---|
| scanTime | scan time |
Partition Scan Metrics¶
The following performance metrics are available only when partitions are used
| Key | Name (in web UI) | Description |
|---|---|---|
| numPartitions | number of partitions read | |
| pruningTime | dynamic partition pruning time |
Dynamic Partition Pruning Scan Metrics¶
The following performance metrics are available only for isDynamicPruningFilter among the partition filters.
| Key | Name (in web UI) | Description |
|---|---|---|
| staticFilesNum | static number of files read | |
| staticFilesSize | static size of files read |
Metadata¶
metadata: Map[String, String]
metadata is part of the DataSourceScanExec abstraction.
metadata...FIXME
inputRDDs¶
inputRDDs(): Seq[RDD[InternalRow]]
inputRDDs is part of the DataSourceScanExec abstraction.
inputRDDs is the single input RDD.
Input RDD¶
inputRDD: RDD[InternalRow]
lazy value
inputRDD is a Scala lazy value which is computed once when accessed and never changes afterwards.
inputRDD is an input RDD that is used when FileSourceScanExec physical operator is requested for inputRDDs and to execute.
When created, inputRDD requests HadoopFsRelation to get the underlying FileFormat that is in turn requested to build a data reader with partition column values appended (with the input parameters from the properties of HadoopFsRelation and pushedDownFilters).
In case the HadoopFsRelation has bucketing specification specified and bucketing support is enabled, inputRDD creates a FileScanRDD with bucketing (with the bucketing specification, the reader, selectedPartitions and the HadoopFsRelation itself). Otherwise, inputRDD createNonBucketedReadRDD.
Creating RDD for Non-Bucketed Read¶
createReadRDD(
readFile: (PartitionedFile) => Iterator[InternalRow],
selectedPartitions: Array[PartitionDirectory],
fsRelation: HadoopFsRelation): RDD[InternalRow]
createReadRDD prints out the following INFO message to the logs (with maxSplitBytes hint and openCostInBytes):
Planning scan with bin packing, max size: [maxSplitBytes] bytes,
open cost is considered as scanning [openCostInBytes] bytes.
createReadRDD determines whether Bucketing is enabled or not (based on spark.sql.sources.bucketing.enabled) for bucket pruning.
Bucket Pruning
Bucket Pruning is an optimization to filter out data files from scanning (based on optionalBucketSet).
With Bucketing disabled or optionalBucketSet undefined, all files are included in scanning.
createReadRDD splits files to be scanned (in the given selectedPartitions), possibly applying bucket pruning (with Bucketing enabled). createReadRDD uses the following:
- isSplitable property of the FileFormat of the HadoopFsRelation
- maxSplitBytes hint
createReadRDD sorts the split files (by length in reverse order).
In the end, creates a FileScanRDD with the following:
| Property | Value |
|---|---|
| readFunction | Input readFile function |
| filePartitions | Partitions |
| readSchema | requiredSchema with partitionSchema of the input HadoopFsRelation |
| metadataColumns | metadataColumns |
Dynamically Selected Partitions¶
dynamicallySelectedPartitions: Array[PartitionDirectory]
lazy value
dynamicallySelectedPartitions is a Scala lazy value which is computed once when accessed and cached afterwards.
dynamicallySelectedPartitions...FIXME
Selected Partitions¶
selectedPartitions: Seq[PartitionDirectory]
lazy value
selectedPartitions is a Scala lazy value which is computed once when accessed and cached afterwards.
selectedPartitions...FIXME
bucketedScan Flag¶
bucketedScan: Boolean
lazy value
selectedPartitions is a Scala lazy value which is computed once when accessed and cached afterwards.
bucketedScan...FIXME
bucketedScan is used when:
- FIXME
Output Data Ordering Requirements¶
outputOrdering: Seq[SortOrder]
outputOrdering is part of the SparkPlan abstraction.
Danger
Review Me
outputOrdering is a SortOrder expression for every sort column in Ascending order only when the following all hold:
- bucketing is enabled
- HadoopFsRelation has a bucketing specification defined
- All the buckets have a single file in it
Otherwise, outputOrdering is simply empty (Nil).
Output Data Partitioning Requirements¶
outputPartitioning: Partitioning
outputPartitioning is part of the SparkPlan abstraction.
Danger
Review Me
outputPartitioning can be one of the following:
-
HashPartitioning (with the bucket column names and the number of buckets of the bucketing specification of the HadoopFsRelation) when bucketing is enabled and the HadoopFsRelation has a bucketing specification defined
-
UnknownPartitioning (with
0partitions) otherwise
Fully-Qualified Class Names of ColumnVectors¶
vectorTypes: Option[Seq[String]]
vectorTypes is part of the SparkPlan abstraction.
Danger
Review Me
vectorTypes simply requests the FileFormat of the HadoopFsRelation for vectorTypes.
doExecuteColumnar¶
doExecuteColumnar(): RDD[ColumnarBatch]
doExecuteColumnar is part of the SparkPlan abstraction.
Executing Physical Operator¶
doExecute(): RDD[InternalRow]
doExecute is part of the SparkPlan abstraction.
Danger
Review Me
doExecute branches off per supportsBatch flag.
Note
supportsBatch flag can be enabled for ParquetFileFormat and OrcFileFormat built-in file formats (under certain conditions).
With supportsBatch flag enabled, doExecute creates a WholeStageCodegenExec physical operator (with the FileSourceScanExec as the child physical operator and codegenStageId as 0) and executes it right after.
With supportsBatch flag disabled, doExecute creates an unsafeRows RDD to scan over which is different per needsUnsafeRowConversion flag.
If needsUnsafeRowConversion flag is on, doExecute takes the input RDD and creates a new RDD by applying a function to each partition (using RDD.mapPartitionsWithIndexInternal):
-
Creates a UnsafeProjection for the schema
-
Initializes the UnsafeProjection
-
Maps over the rows in a partition iterator using the
UnsafeProjectionprojection
Otherwise, doExecute simply takes the input RDD as the unsafeRows RDD (with no changes).
doExecute takes the numOutputRows metric and creates a new RDD by mapping every element in the unsafeRows and incrementing the numOutputRows metric.
Tip
Use RDD.toDebugString to review the RDD lineage and "reverse-engineer" the values of the supportsBatch and needsUnsafeRowConversion flags given the number of RDDs.
With supportsBatch off and needsUnsafeRowConversion on you should see two more RDDs in the RDD lineage.
Creating FileScanRDD with Bucketing Support¶
createBucketedReadRDD(
bucketSpec: BucketSpec,
readFile: (PartitionedFile) => Iterator[InternalRow],
selectedPartitions: Array[PartitionDirectory],
fsRelation: HadoopFsRelation): RDD[InternalRow]
Danger
Review Me
createBucketedReadRDD prints the following INFO message to the logs:
Planning with [numBuckets] buckets
createBucketedReadRDD maps the available files of the input selectedPartitions into PartitionedFiles. For every file, createBucketedReadRDD getBlockLocations and getBlockHosts.
createBucketedReadRDD then groups the PartitionedFiles by bucket ID.
Note
Bucket ID is of the format _0000n, i.e. the bucket ID prefixed with up to four 0s.
createBucketedReadRDD prunes (filters out) the bucket files for the bucket IDs that are not listed in the bucket IDs for bucket pruning.
createBucketedReadRDD creates a FilePartition (file block) for every bucket ID and the (pruned) bucket PartitionedFiles.
In the end, createBucketedReadRDD creates a FileScanRDD (with the input readFile for the read function and the file blocks (FilePartitions) for every bucket ID for partitions)
Tip
Use RDD.toDebugString to see FileScanRDD in the RDD execution plan (aka RDD lineage).
// Create a bucketed table
spark.range(8).write.bucketBy(4, "id").saveAsTable("b1")
scala> sql("desc extended b1").where($"col_name" like "%Bucket%").show
+--------------+---------+-------+
| col_name|data_type|comment|
+--------------+---------+-------+
| Num Buckets| 4| |
|Bucket Columns| [`id`]| |
+--------------+---------+-------+
val bucketedTable = spark.table("b1")
val lineage = bucketedTable.queryExecution.toRdd.toDebugString
scala> println(lineage)
(4) MapPartitionsRDD[26] at toRdd at <console>:26 []
| FileScanRDD[25] at toRdd at <console>:26 []
createBucketedReadRDD is used when:
FileSourceScanExecphysical operator is requested for the input RDD (and the optional bucketing specification of the HadoopFsRelation is defined and bucketing is enabled)
needsUnsafeRowConversion Flag¶
needsUnsafeRowConversion: Boolean
needsUnsafeRowConversion is enabled (i.e. true) when the following conditions all hold:
-
FileFormat of the HadoopFsRelation is ParquetFileFormat
-
spark.sql.parquet.enableVectorizedReader configuration property is enabled
Otherwise, needsUnsafeRowConversion is disabled (i.e. false).
needsUnsafeRowConversion is used when:
FileSourceScanExecis executed (and supportsBatch flag is off)
supportsColumnar Flag¶
supportsColumnar: Boolean
supportsColumnar is part of the SparkPlan abstraction.
supportsColumnar...FIXME
Demo¶
// Create a bucketed data source table
// It is one of the most complex examples of a LogicalRelation with a HadoopFsRelation
val tableName = "bucketed_4_id"
spark
.range(100)
.withColumn("part", $"id" % 2)
.write
.partitionBy("part")
.bucketBy(4, "id")
.sortBy("id")
.mode("overwrite")
.saveAsTable(tableName)
val q = spark.table(tableName)
val sparkPlan = q.queryExecution.executedPlan
scala> :type sparkPlan
org.apache.spark.sql.execution.SparkPlan
scala> println(sparkPlan.numberedTreeString)
00 *(1) FileScan parquet default.bucketed_4_id[id#7L,part#8L] Batched: true, Format: Parquet, Location: CatalogFileIndex[file:/Users/jacek/dev/oss/spark/spark-warehouse/bucketed_4_id], PartitionCount: 2, PartitionFilters: [], PushedFilters: [], ReadSchema: struct<id:bigint>, SelectedBucketsCount: 4 out of 4
import org.apache.spark.sql.execution.FileSourceScanExec
val scan = sparkPlan.collectFirst { case exec: FileSourceScanExec => exec }.get
scala> :type scan
org.apache.spark.sql.execution.FileSourceScanExec
scala> scan.metadata.toSeq.sortBy(_._1).map { case (k, v) => s"$k -> $v" }.foreach(println)
Batched -> true
Format -> Parquet
Location -> CatalogFileIndex[file:/Users/jacek/dev/oss/spark/spark-warehouse/bucketed_4_id]
PartitionCount -> 2
PartitionFilters -> []
PushedFilters -> []
ReadSchema -> struct<id:bigint>
SelectedBucketsCount -> 4 out of 4
As a DataSourceScanExec, FileSourceScanExec uses Scan for the prefix of the node name.
val fileScanExec: FileSourceScanExec = ... // see the example earlier
assert(fileScanExec.nodeName startsWith "Scan")
When executed, FileSourceScanExec operator creates a FileScanRDD (for bucketed and non-bucketed reads).
scala> :type scan
org.apache.spark.sql.execution.FileSourceScanExec
val rdd = scan.execute
scala> println(rdd.toDebugString)
(6) MapPartitionsRDD[7] at execute at <console>:28 []
| FileScanRDD[2] at execute at <console>:27 []
import org.apache.spark.sql.execution.datasources.FileScanRDD
assert(rdd.dependencies.head.rdd.isInstanceOf[FileScanRDD])
FileSourceScanExec supports bucket pruning so it only scans the bucket files required for a query.
scala> :type scan
org.apache.spark.sql.execution.FileSourceScanExec
import org.apache.spark.sql.execution.datasources.FileScanRDD
val rdd = scan.inputRDDs.head.asInstanceOf[FileScanRDD]
import org.apache.spark.sql.execution.datasources.FilePartition
val bucketFiles = for {
FilePartition(bucketId, files) <- rdd.filePartitions
f <- files
} yield s"Bucket $bucketId => $f"
scala> println(bucketFiles.size)
51
scala> bucketFiles.foreach(println)
Bucket 0 => path: file:///Users/jacek/dev/oss/spark/spark-warehouse/bucketed_4_id/part=0/part-00004-5301d371-01c3-47d4-bb6b-76c3c94f3699_00000.c000.snappy.parquet, range: 0-423, partition values: [0]
Bucket 0 => path: file:///Users/jacek/dev/oss/spark/spark-warehouse/bucketed_4_id/part=0/part-00001-5301d371-01c3-47d4-bb6b-76c3c94f3699_00000.c000.snappy.parquet, range: 0-423, partition values: [0]
...
Bucket 3 => path: file:///Users/jacek/dev/oss/spark/spark-warehouse/bucketed_4_id/part=1/part-00005-5301d371-01c3-47d4-bb6b-76c3c94f3699_00003.c000.snappy.parquet, range: 0-423, partition values: [1]
Bucket 3 => path: file:///Users/jacek/dev/oss/spark/spark-warehouse/bucketed_4_id/part=1/part-00000-5301d371-01c3-47d4-bb6b-76c3c94f3699_00003.c000.snappy.parquet, range: 0-431, partition values: [1]
Bucket 3 => path: file:///Users/jacek/dev/oss/spark/spark-warehouse/bucketed_4_id/part=1/part-00007-5301d371-01c3-47d4-bb6b-76c3c94f3699_00003.c000.snappy.parquet, range: 0-423, partition values: [1]
FileSourceScanExec uses a HashPartitioning or the default UnknownPartitioning as the output partitioning scheme.
FileSourceScanExec supports data source filters that are printed out to the console (at INFO logging level) and available as metadata (e.g. in web UI or explain).
Pushed Filters: [pushedDownFilters]
Logging¶
Enable ALL logging level for org.apache.spark.sql.execution.FileSourceScanExec logger to see what happens inside.
Add the following line to conf/log4j2.properties:
log4j.logger.org.apache.spark.sql.execution.FileSourceScanExec=ALL
Refer to Logging.